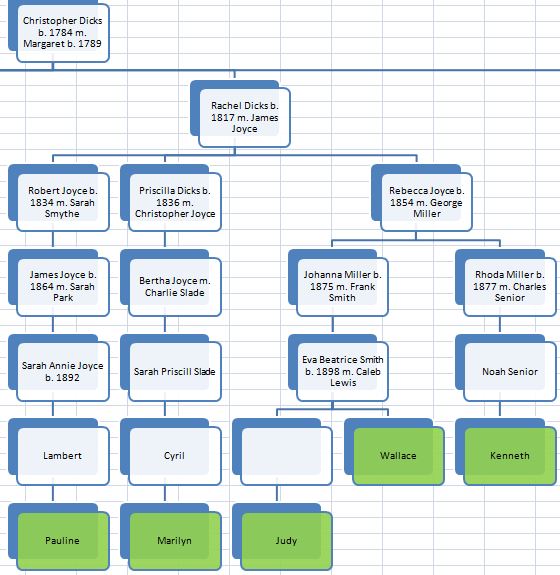
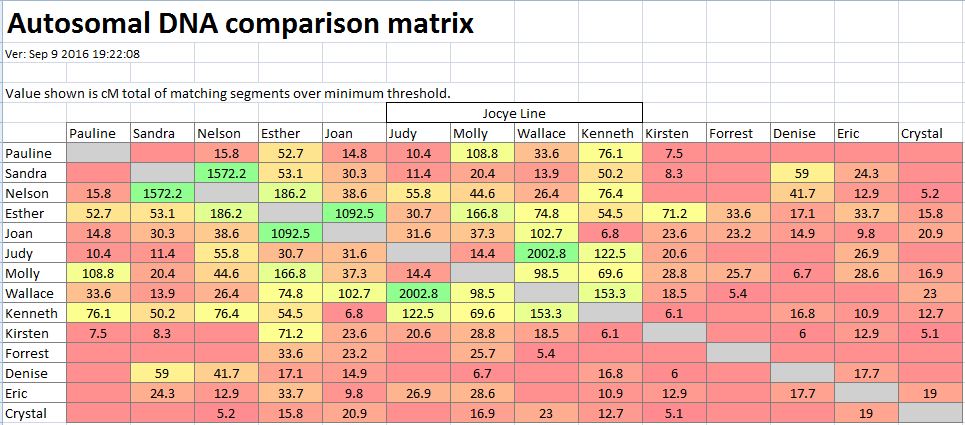
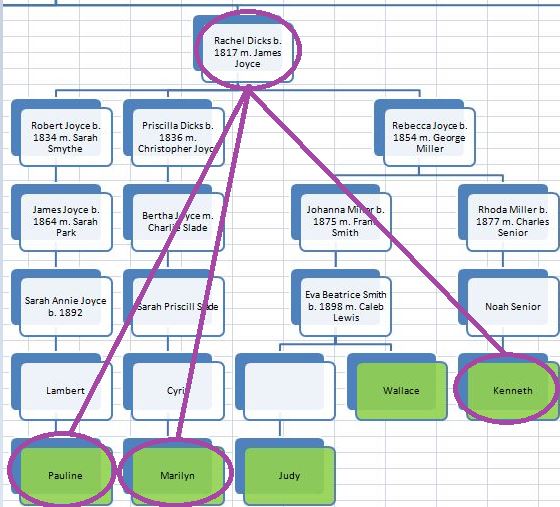
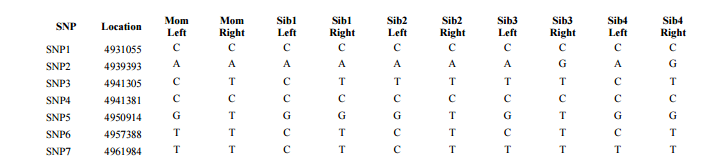
In my last Blog on raw data phasing, I went through 3 principals that Whit Athey laid out in a paper on phasing raw data when one parent’s DNA results were missing. Using those principals, and the MS Access program, I was able to sort many of my bases and 2 sisters’ bases into ones we received from our mom and ones that we received from our dad. I checked a few of my results with a chromosome map made for me by M Macneill.
Paternal Patterns
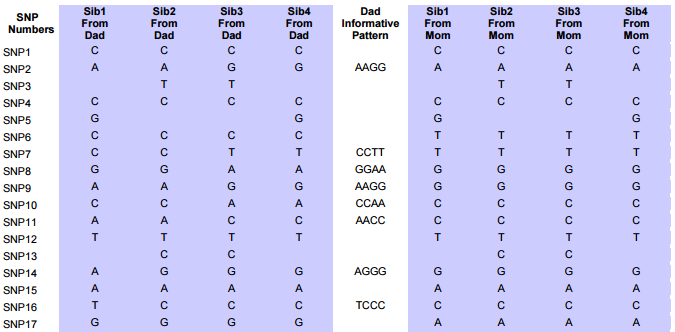
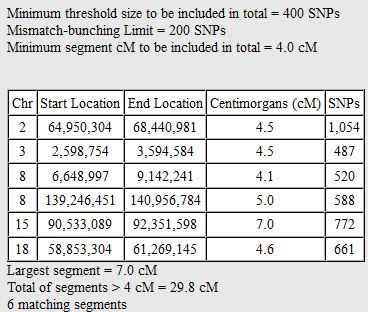
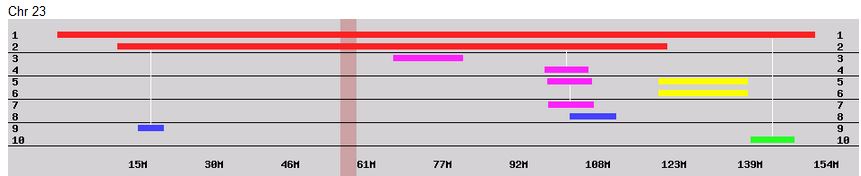
I had gotten to the part of the Athey paper where he talks about paternal patterns of bases that the sibling combinations received. I noted a space between the first two paternal patterns that I looked at. Below the pattern goes from an ABA pattern to an ABB pattern.

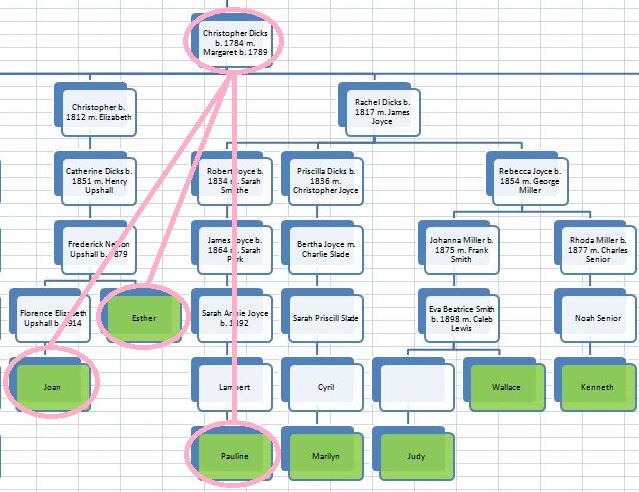
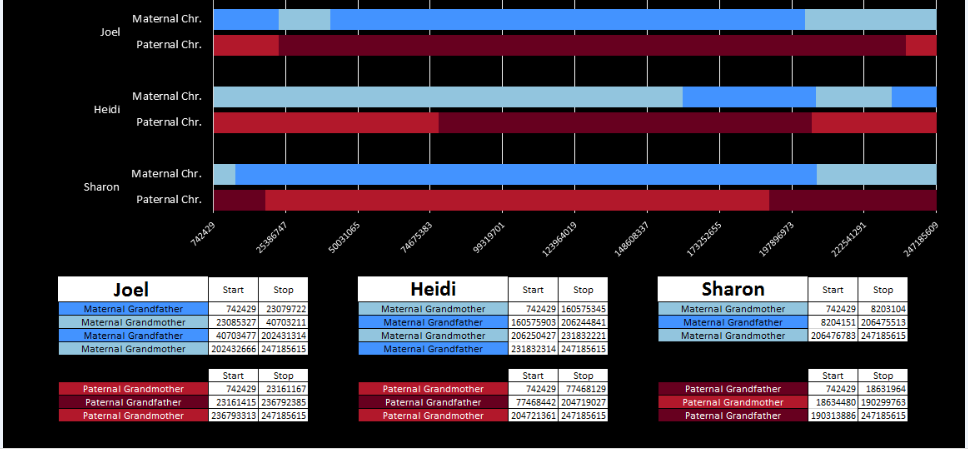
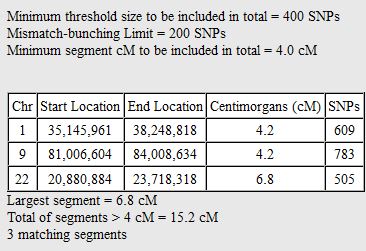
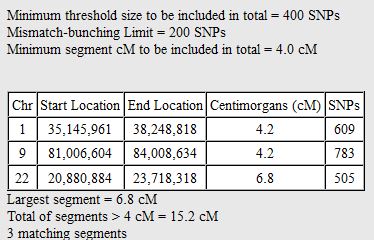
There was a gap between the ABA and ABB pattern where there was no ‘pattern’ as my 2 sisters and I shared the same base there. When my sisters and I all share the same base, that is an AAA “pattern”. That AAA area corresponded exactly to the area between the 2 yellow lines below in the chromosome map made for me by M MacNeill – prairielad_genealogy@hotmail.com .
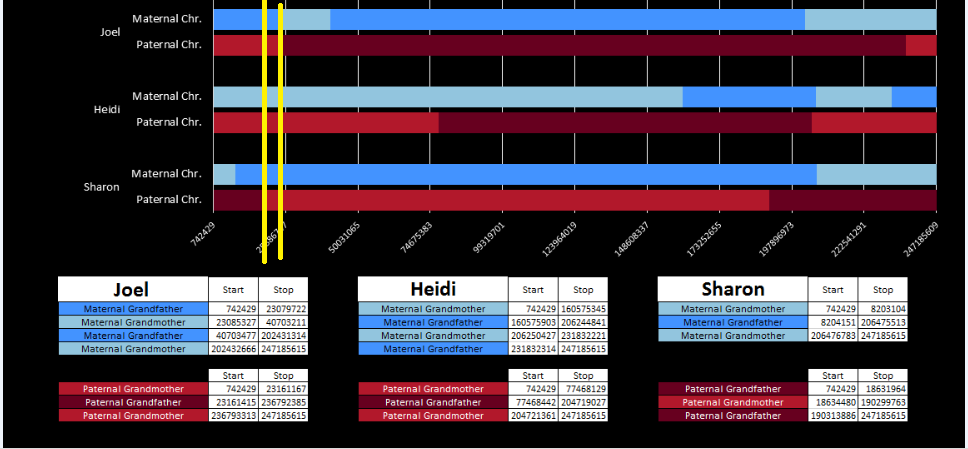
In the map above, MacNeill was able to determine that my 2 sisters and I got our DNA from our paternal grandmother in the area between the 2 yellow lines. Further, the first yellow line described Sharon’s first paternal crossover point and the second yellow line described my (Joel’s) first paternal crossover point.
Finding All the Paternal Crossover Points
At this point in the Athey Paper, he recommended looking at the paternal pattern and filling in the missing bases based on the known pattern. I was looking for an easier way to do this, so decided to take a different approach. I decided that I would find all the paternal crossover points first. Then, armed with that information, I would create a formula that would fill in most or all of the missing bases for each pattern.
However, this required a modification of my database to make the work easier. I wanted a number to define the range of patterns, so that I could apply an easy query to add missing bases. I already had this but I hadn’t used it. Back when I imported the 4 sets of raw data into Access, Access assigned an ID to every row of data. That meant that I needed to add that ID into all the queries that I had done previously to make tables and further queries. This took a while, but I believe that it was worth it.

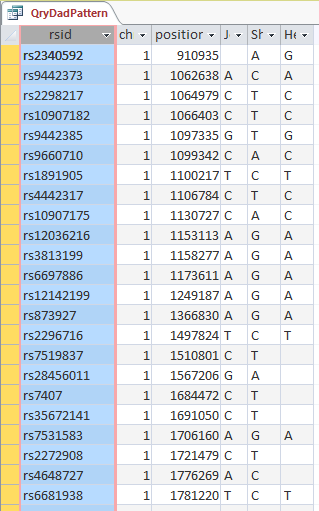
The ID is the first column.
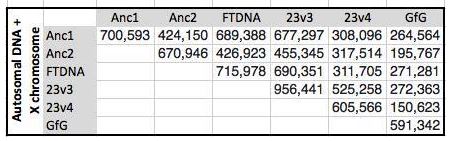
I started going down all my data and noting the change of each pattern. I put the results into an Excel table. Here the Start and Stop numbers are the Access assigned ID numbers. The ID’s corrrespond with the number of DNA locations looked at. In this case there were a bit over a total of 700,000 of these locations for my mom, my 2 sisters, and me.
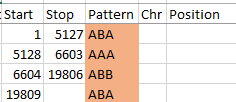
Then I noted the patterns are repeating as would be expected. For example, my first pattern was ABA, but 3 patterns later, that same ABA repeated. My thought was to create a query just for ABA patterns. Then when scrolling down looking for changes, the separation between rows should be greater and it would be easier to see where those changes were.
Here is what my Access query looks like. I changed the query name to DadSpecificPattern.
 query
query
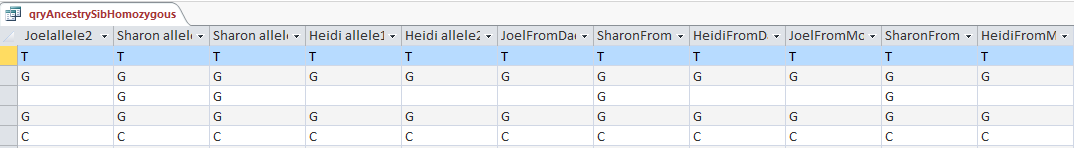
This particular query gives me the ABB pattern. I have the HeidifromDad base equal to the SharonFromDad base. That makes me the A and Sharon and Heidi the BB of the ABB Pattern. If you think about it, that also means in these areas that Heidi and Sharon will have their base from the same paternal grandparent and mine will be from the other paternal grandparent. I’m learning as I go. I’m sure that information will come in handy later.
My plan seemed to be good, but there was one catch. Once I refined my query, most or all of the blanks disappeared. That meant that the start and end points might not be exact. Here is an example of what I mean.
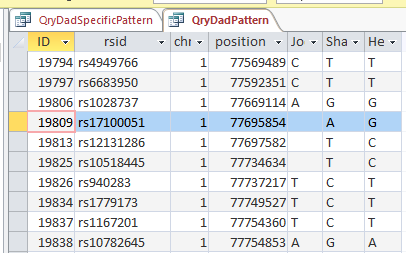
This is from my old Dad Pattern query with the blanks still there. The change from ABB to ABA happens at ID or line 19809. However, the new query takes out the blanks to make it look like the change is at ID Line 19826.
Here is what my DNA results look like so far without a filter (or query). The last 3 columns are the bases from Dad columns. There is a lot going on between lines 19809 and 19826.
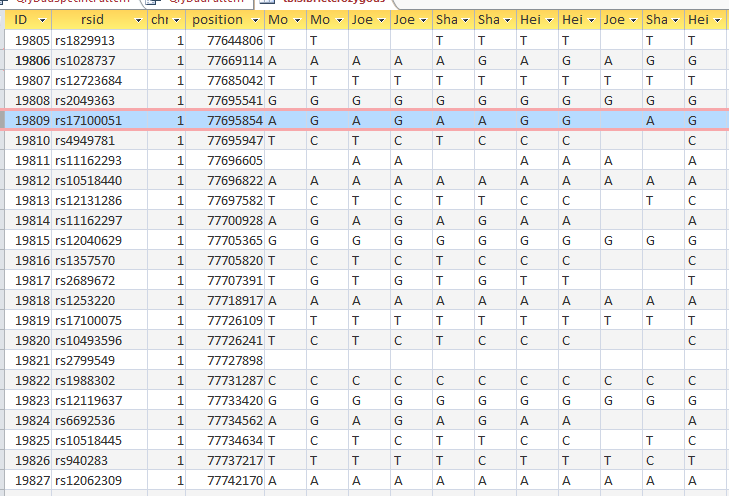
Once I apply a formula to add bases, it will say something like: In the lines that have the ABA pattern where there is a blank at either A spot, replace the blank with the A that is there. If I apply the rule too late, I will be missing an area. Worse, If I were to use the 19826 cutoff, I may be still using the previous rule. That rule would say basically the same thing except, “Where the row is ABB and one of the B’s is missing replace the missing B with the one that is there.” If I apply an ABB rule to an ABA area, I’ll get bad results.
Long story short, I ended up recording a rough start and stop in my Excel Spreadsheet.

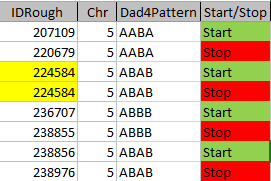
I started naming the segments, but realized that was not necessary. Some of the patterns were only at one point rather than in a long segment. I believe that is an anomaly due to a bad read, mutation or some other problem. Those are the ones in the spreadsheet that had no end point. It took me part of a morning to get all the paternal crossover pattern points for all 23 chromosomes. Fortunately for 3 siblings, the patterns are only ABA, AAB and ABB.
I just went back and checked the error points/aonomalies. I reran the Heterozygous Sibling Query and it fixed at least the first problem and hopefully the others. When I added the ID’s in, I had to redo all the queries quickly, so I suppose that is where the errors came in. That is not a problem as long as the problem can be found a fix can usually also be found. There actually weren’t that many errors. There are still some anomalies that are just anomalies. I have left those in yellow in the spreadsheet image below.
So in my spreadsheet, I have all the rough starts and ends for all the crossovers for my 2 sisters and myself. Here is the top part of the spreadsheet sorted by rough start:

Next, all I need are more exact start and end points. Here is the start of what I have:
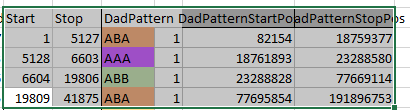
I picked this section because it looks pretty complete already. Note that my Start and Stop numbers are pretty close to each other. That means that there are no other AAA segments in-between. I had to do an additional Access query to add in the position numbers for the Start and Stop of each chromosome’s pattern change. This was important if I want to convert the results from Build 37 to Build 36 to compare to MacNeill’s work or to gedmatch.com.
Starting to Find Paternal Crossovers and Assigning to Siblings
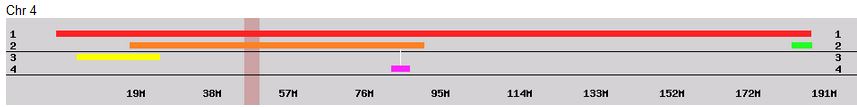
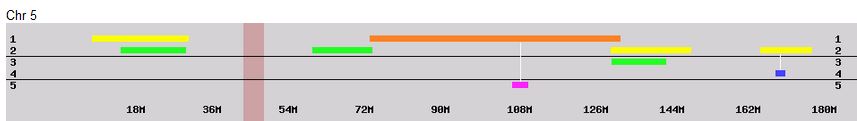
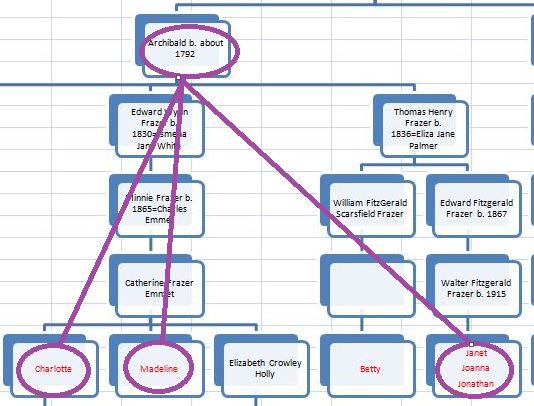
Previously I had been calling the start and end of my patterns crossovers. These two terms aren’t totally interchangeable as the start or stop of a pattern may happen at the beginning or end of a Chromosome and therefor not be a crossover at that point. It seems like it should be pretty easy to find the crossovers. Look at the image above. The first and second rows show ABA going to AAA. The order in me and my siblings are JSH or Joel, Sharon and Heidi. The only letter that changes is the B to A. That is the position that Sharon is in, so the paternal crossover has to go to her. From row 2 to row 3 the pattern changes from AAA to ABB at Chromosome 1, position 23,288,828, Build 37. That doesn’t mean that 2 siblings have a crossover there as we are looking at the patterns, not the letters. It is actually the letter that stayed the same that represents the crossover here. AAA to ABB means: all the same (AAA) goes to one different and 2 the same (ABB) – in this case Sharon and Heidi). The one that is different is me and I get the crossover at this location. The next change is from ABB to ABA. This is a little harder to see. I would say that that this crossover goes to Heidi if my reasoning is right. BB was the same before and goes to BA. It must be Heidi that changed because now she matches Joel who didn’t change. I’ll need to figure out how to make better bar graphs in Excel, but here is how the beginning part my father’s Chromosome 1 broke up for 3 of his children. Or another way to look at it the vertical lines are where my father’s maternal and paternal chromosomes combined in each of his 3 children that we are now looking at.
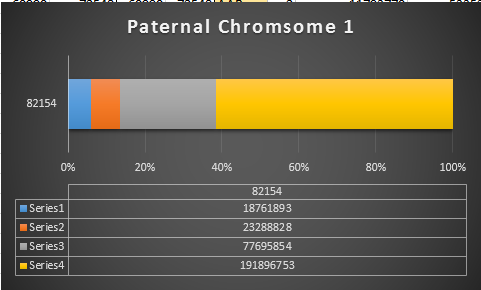
Where:
- Series 1 is Sharon. Where the color goes from blue to orange is where Sharon has a change from one paternal grandparent’s DNA to another paternal grandparent’s DNA. The number to the right of Series 1 is the Build 37 Chromosome position number for Sharon’s crossover.
- Series 2 is Joel’s first crossover (between orange and gray) and
- Series 3 is Heidi’s first crossover position between gray and yellow [The same explanation under Sharon above applies to Joel and Heidi]
I’ll go back to the M MacNeill Standard. It’s like having an answer sheet to my questions.

According to MacNeill, I have assigned the crossovers to the correct siblings. In the above chart, just look at the red. I haven’t gotten to the maternal part yet, which MacNeill has in blue. The first 3 crossovers are where the red changes from light to dark or dark to light red. The difference in the MacNeill Chart is that his chart is split out one bar for each sibling. The other difference is that MacNeill has build 36 Chromosome position numbers and the numbers I have are from Build 37.
The Process
- Phase the siblings into maternal and paternal DNA using the principles that Athey outlines
- Find the paternal and maternal crossovers by pattern changes
- Assign the crossovers to the correct sibling using the pattern changes
- Assign the segments to the correct grandparent. This requires knowledge of cousin matches on the appropriate grandparent side.
That is the big picture which I am understanding as long as I don’t get too lost in the details.
Back to the Details: Fill in More A’s, G’s, C’s and T’s
I have been setting up my data for this, so hopefully, this will be easy. I now have 3 areas to look at:
AAB paternal update
Now I go back to my spreadsheet and sort it by Dad Pattern:

The Start and Stop areas are the ones I want to update. First, I’ll copy my most up to date Table in Access which is tblSibHetorzygous. I’ll rename that tblDadPatternUpdate. Then I want to look for missing data and update the blanks using the AAB pattern.
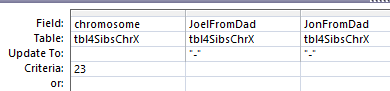
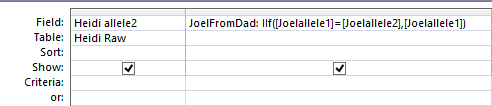
In Access, I create a query with the new table.

I chose the position fields and Paternal Pattern fields. I will change this to an update query which adds an Update To row. The criteria I want is when JoelFromDad = Sharon from Dad (AAB). Actually, I forgot, I was going to use ID criteria. So in the ID field, I need a lot of information. For the first AAB segment, I need everything between ID 45393 and 54155. This is what the criteria looks like:

When I choose that area, I get over 8,000 lines. However, I only want to update when there is one missing value in the first 2 and the one that isn’t missing is not equal to the third. Here is the result of the above query in my first AAB area:

I assume that the first blank should be a T. This would be one of the AAA results by chance in an AAB area. I don’t want to fill in the second line as I don’t know if it will be GGG or something else. That is what I meant by saying I don’t want to fill anything in unless there is only one missing value. In the 5th line there is A?G. That would have to be AAG (in an AAB Pattern area). There are some lines that have everything missing that I don’t want to touch.
How to create a query?
First, I want the situation where Joel doesn’t equal Sharon or Joel Doesn’t equal Sharon. That would create an AAB situation:

This query results in 1,666 rows of data including rows that are already filled in. Note that I had to write the range of ID’s twice because in order to get an OR situation I needed to put Joel not equat to Heidi and Sharon not equal to Heidi on separate lines. A simpler query is this one:
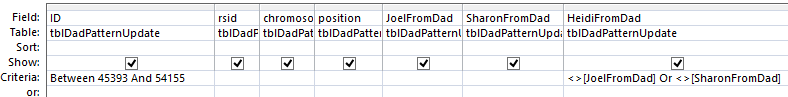
The above achieves the same results in one line. Now, for this query, if Joel is blank, replace it with Sharon’s results. If Sharon is blank, replace it with Joel’s results. Here is the query prior to the updating part:

This shows that there are 29 blanks for Joel and Sharon meeting this AAB criteria in the first range of AAB’s:
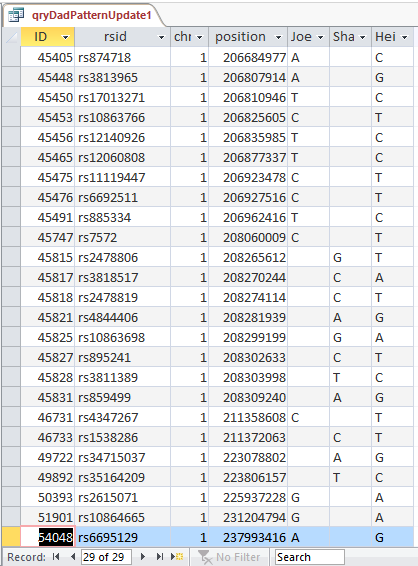
Next, I apply the same logic to all the AAB segments. In the Expression Builder of Access, I type in this simple formula:
Between 45393 And 54155 Or Between 60990 And 72548 Or Between 207109 And 220679 Or Between 313271 And 317516 OR Between 326845 And 326912 OR Between 389395 And 390311 OR Between 400045 And 405578 OR Between 419982 and 427158 OR Between 433191 And 446672
OR Between 482297 And 492542 OR Between 532520 And 539292 OR Between 571557 And 579594 OR Between 589614 And 589666 OR Between 630037 And 630314 OR Between 630319 And 630378 OR Between 658744 And 659375 OR Between 670533 And 672360 OR Between 673325 And 682544
Simple but long. This has the AAB Starts and Stops for 23 chromosomes. Then I copy it into the next ID criteria line and get this result:

It took a few minutes to type the criteria, but the goal is to update 1,514 lines of missing Paterrnal Pattern data with the push of one button. I still think it is quicker than going line by line and will be more accurate if I got the criteria right.
Next, I change the above Select Query to an Update Query.

When my (Joel’s) base from Dad is missing, I update to Sharon’s base. When Sharon’s base from Dad is missing her base is updated with mine. Isn’t sharing great? I didn’t look at the case where Heidi’s base from dad was missing, because if that was missing we wouldn’t be able to see any AAB Pattern.
Let’s UPdate
I push the run button and check the results. Here is my standard dire warning:

Now I will check if it worked. I’ll try ID or Line # 682124:

Unfortunately, that was an undesirable result. Before I had A?G. I changed this to ?AG. It appears that my query both replaced my value with Sharon’s, but replaced Sharon’s with my blank. I hadn’t expected that. Next, I’ll check ID# 682182. I had ?AG and replaced it with A?G. So until, I can think of a solution, I’ll need to split the 2 queries.
Fix it! Quick!
First I recopied by Heterozygous Sibling Table back to the Dad Pattern Update 1 Table. This got the table back to the way it was. Here is my simpler query.

Here if my base from Dad is null, replace it with Sharon’s base from Dad. I’ll check ID# 682182 again:

This gets into the category of trial and error. Sharon’s result still got replaced with nothing. See in the previous query I still was telling Access to put update Sharon’s results with mine. I needed to take that out:

There. Now the SharonFromDad Update To is blank. I go through the same procedures and now it looks right.

We now went from ?AG to AAG in the last 3 columns. These are the bases from Dad columns.
The next step is pretty easy:

I took out my criteria and put criteria in the SharonFromDad field. When she has a blank, replace it with Joel’s base from Dad. I hit run and it updated over 600 rows. Here is my original check spot at ID# 682124 with better results in the last 3 columns:
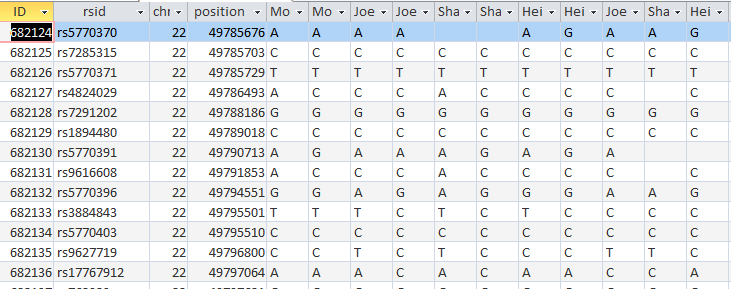
It took a while, but at least I got it right. The moral of the story is to not ask Access to do 2 things at once when those 2 things involve the same 2 people.
The Next Step: ABA
This time I’ll try a different query. I want there to be a B from the ABA in each case, so I’ll make sure that Sharon’s base from Dad is there:

Maybe I’ll figure what went wrong last time or come up with a new error. Above, I want the criteria on the first line to be for my blank base: If Sharon’s base from Dad is not equal to Heidi’s Base from Dad Put Heidi’s base from Dad in my blank spot. For Heidi, When Joel’s base from Dad doesn’t equal Sharon’s base from Dad, put Joel’s Base in Heidi’s spot.
I’m so tempted to try this query, but before I do, I’ll copy the previous table of the DadPatternUpdate to a new Dad Pattern Update ABA Table. This will preserve what I have in the now older DadPatternUpdate Table in case anything goes wrong. Hey, what could go wrong?

I pushed the Update Button and updated over 30,000 rows. The results don’t appear to be any better, so I’m back to my 2 step process.
Here is my new slimmed down query:
This new Update Query should update my Line 18 in the new UpdateABA Dad Pattern Table and it does:
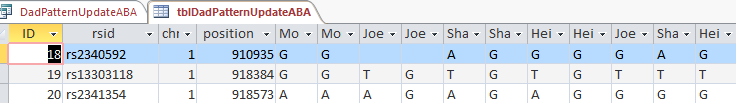
I now have a full ABA pattern on that line. According to Access over 30,000 Lines were updated, so it wasn’t a total waste of time.

Run and check Line 149:

We have ABA in the last 3 columns, so that is good. Line 18 is still OK. I checked it just to make sure.
Query AAB Revised
After seeing how well the ABA Query went, I decided to revise the old AAB Query:

This is now looking at over 37,000 rows. This updates my AAB Blanks to tblDadPatternAAB. I don’t know if it is a better query, but at least I’m being consistent.

This was over 80,000 rows, so I’ll assume that bigger is better.
I copied that resulting Table to tblDadPatternUpdateABA and reran the 2 ABA Update Queries. Here is one of the rerun queries updating the ABA Paternal Table:
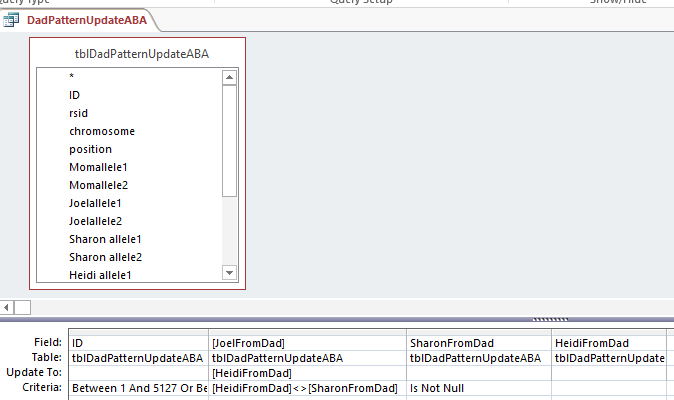
Down to ABB
My Last updated Paternal Table was updating ABA, so I’ll copy that to a new Table called tblDadPatternUpdateABB. I’ll also copy my last query and put in the appropriate Starts and Stops for the paternal ABB patterns. Again,
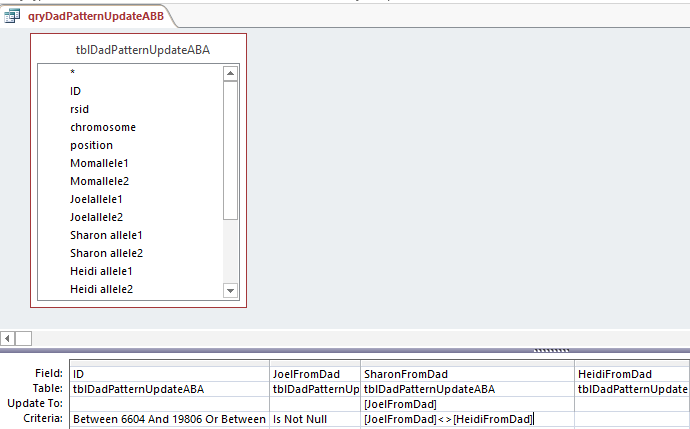
This says when Joel’s base from dad is not the same as Heidi, put that Joel from Dad into the space. Probably a more precise query would have said when Sharon from Dad is null and Joel from Dad is not equal to Heidi from Dad. I suppose technically the above query could be writing over a base with the same base in most cases.
I’ll fix that and notice that I had the wrong table in the top, so I’ll change that also.

This only updated 944 rows, so maybe bigger is not better. Here is Part 2:

This was almost 3,000 rows updated. Now I should check if it worked. I scrolled for an ABB Pattern in an old query and found this:

Here is my check:

I guess I’ve been working too long. Here I have an AAB instead of the ABB I wanted. That is because I had Heidi updated to me (the A) instead of Sharon (the B). Here is the correction:

I made a fresh Table of ABB. When I opened up the Query, it was saved this way:
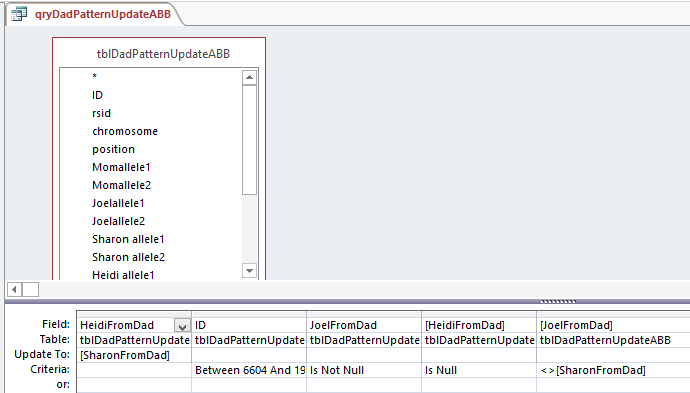
So Access changed my query. Note that there are 2 fields with HeidiFromDad in them. One is for the Update To and the other has Criteria. That is probably a clearer way to do it. Who should argue with Access?
I updated that and I take a cue from Access for Part 2:

In English, the above says, “For this range when JoelFromDad is not blank but Sharon from Dad is, and Joel from Dad has a different value that Heidi from Dad, put that Heidi from Dad value where Sharon had the blank. It sounds a little complicated.
Back to Row 197704 and I’ll look at 197709 while I’m at it:

Oh no, it is still wrong! I checked the previous ABA Table and that was the reason for the error. The error is also in the old AAB Table. However, the error was not in the file before that. My guess is that the AAB rule got applied to the wrong range of rows. I don’t see an error there, so I’ll have to rerun all the queries.
That’s OK, because I’m brushing up on the queries and will use the Is Null value so we will only be filling in the missing bases.

I had more problems, so I deleted the AAB Table and recopied the previous Table into it. I reran the Revised AAB Query halfway and it looked OK. However, when I ran the second half of the AAB query – filling Sharon’s results, the problem came back at ID# 197704. Very mysterious. The problem was where I thought it was originally. Look at the ID Criteria for the AAB Pattern Query:
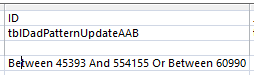
There is an extra digit in the first between. The range goes from 45393 to 544155. The second number should be 54155. So this query was performed on 450,000 more rows than intended. I updated the AAB query with fewer rows. Again fewer is better. After many requeryings, I got the desired result for ID# 197704:

That should be the end of the first phase of nit picky work on the Paternal Side.
Summary, Conclusion and What’s Next
- This was a lot of work, but the good news is that this update is for all the Chromosomes at once.
- The bad news is that I have to do this again for the Maternal Side
- Next up should be easy. That is just re-applying the Principles that Whit Athey Outlined on the new bases that I added from knowing the patterns. This should update missing maternally received bases from the updated paternally received bases.
- I haven’t filled in blanks for the AAA patterns yet.
- I am a little ahead of the game as I looked at how some of the first paternal crossovers will look.
- Also with some basic phasing, I was able to deduce who those first paternal crossovers belonged to – one each to my two sisters and one for me.
- If anything can go wrong it will