Mom Patterns
Up to this point, I have phased 4 siblings based on 3 principles outlined by Whit Athey. I have looked at the bases the 4 siblings had from their Dad. Those Dad bases made up patterns. Based on those patterns, other Dad bases were added to those siblings within those pattern areas. After those bases were added, mom bases were added where the siblings were heterozygous. The changes were documented in a Base Tracker.
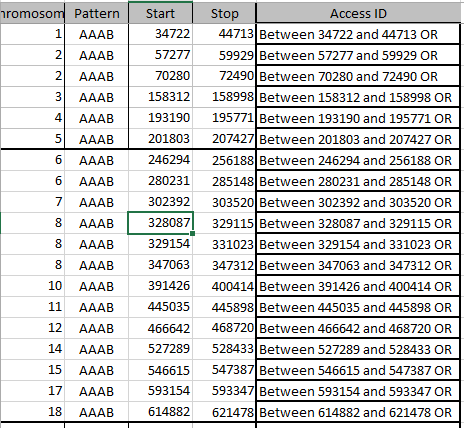
Start stop using access min max – AAAB Mom Pattern
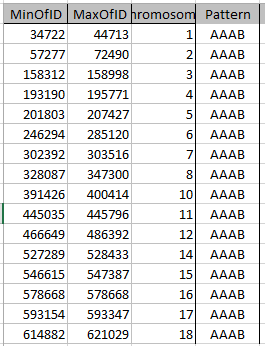
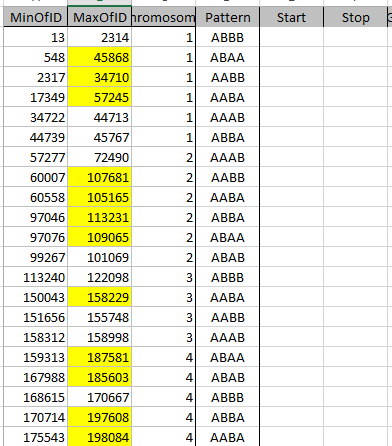
I can just look at my previous Blog to see what I did for my dad pattern. The results of this query:
get copied to this spreadsheet where I added a column for Pattern:
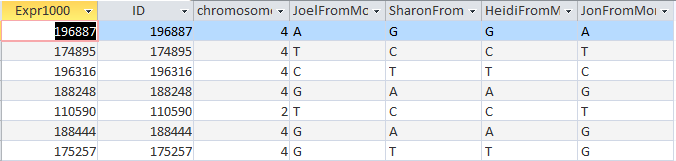
That was my big time saving step from my last query. Before I run each Min Max Total Query, I check a regular Select Query to make sure I have the right pattern. For example, here is my ABBA Mom Pattern check:
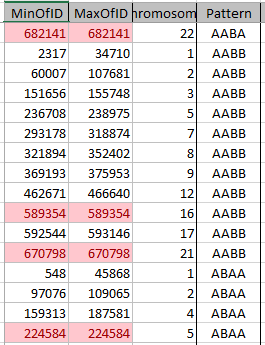
In a few minutes, I have 111 Start/Stop Mom Pattern pairs. This time, I’ll add conditional formatting to point out the one position patterns:
These single patterns tend to mess me up as I’m looking for patterns, so I’ll take them out of my spreadsheet, but not out of my Access data tables. There were 10 of these. I don’t know if that is a lot.
Getting better starts and stop for the mom patterns
The next step takes a little while. I look at the [now] 95 Start/Stop pairs for the various patterns. I highlighted the overlapping areas in yellow:
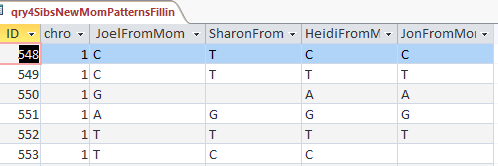
Actually, the first pattern overlaps into the second also. Some of these may be caused by single location patterns. For example at Chromosome 1, when I got to ID# 548 I find this:
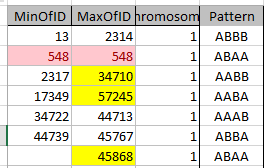
There is an ABAA Pattern, but it only lasts for one position and then is on to an ABBB pattern. I copied the end location for ABAA and put it at the end of Chromosome 1 to check later and made note of the one position pattern:
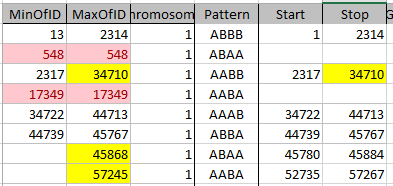
After that, it makes more sense that the ABBB pattern Stop at 2314 goes into an AABB Pattern Start at 2317. Here is the adjusted Chromosome 1 for my siblings’ Mom Patterns:
I moved the first Start to the Start of Chromosome 1 and last Stop to the end of Chromosome 1 as they were already pretty close to those positions. All combinations of patterns are represented here except for ABAB. I don’t have a start and stop for the single patterns as I’ll be taking them out later.
Filling In Mom Patterns
Now that I have all the mom patterns and their starts and stops as well as I can, I will fill in the patterns. I’ll start with AAAB. First I use the Concatenate formula in Excel to get my starts and stops in Access language. Then I sort the patterns in Excel:
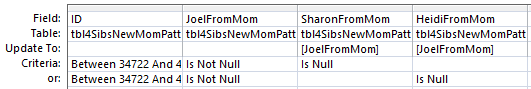
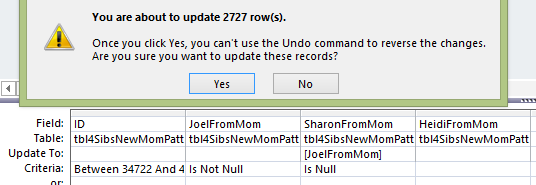
I have 19 AAAB Mom Patterns. Next I go into Access and create an Update Query using the table called tbl4SibsNewMomPatternsFillin. In the AAAB Pattern, I will want to fill in the missing A’s.
This looks like a good query, but I want to track how many bases I’m updating, so this query would make it difficult to track that as I’m adding bases to Sharon and Heidi. So again, I will go with the simpler query.
Here is the first Mom Pattern Fill-in update on the Base Tracker:
I continued the same process down the Mom Patterns, filling in what was missing from each of the siblngs:
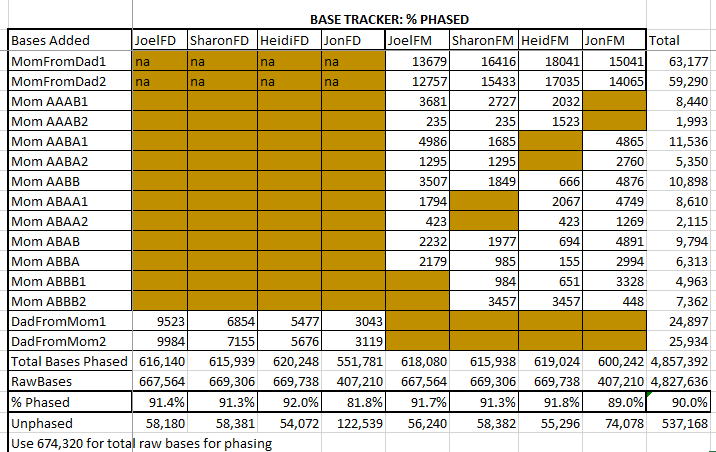
In each case for each pattern, I added less than 5,000 bases to each sibling. I also added to my spreadsheet a percentage of overall phasing which is now at 89.1%. This is how the 4 siblings are phased on average. Jon, who tested with the Ancestry V2 is bringing the other siblings’ overall average down.
Principle 3 – Dad Bases From Mom Bases
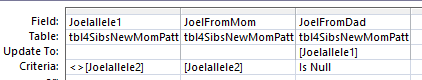
This is the icing on the cake for me. After all the work of determining Patterns and Starts and Stops, I have an easy step to add bases. Principle 3 says if you are heterozygous and you know one of your bases is assigned to one parent, then the other base must be assigned to the other parent.
I had to look at my previous blog to see how I did this. Let’s see if this looks right:
The first column makes sure that I am heterozygous as my 2 alleles are not the same. The 2nd columns says that I know that I got allele2 from Mom. The 3rd column says to put my allele1 as the one I got from dad. That seems to make sense. This results in 9523 rows of updates in 22 Chromosomes. In part 2 of this Update Query, I switch the alleles:
This says if my allele1 is from Mom assign allele2 to be from Dad.
Summary of Pattern Filling In and Dad Bases from Mom Bases
Here the overall phasing is 90%, but I had a pretty strict measure of phasing. It involved alleles that Jon was not even tested for. Here we are getting a diminishing return. I could continue the process, but I won’t.
Next Steps
Now I have a good idea where all the crossovers are. I need to assign those to siblings. Then I need to figure out how to portray the final results.
Assigning Crossovers to Siblings
I might as well jump right in. I’ll try a Chromosome that McNeill has mapped. Actually, he only did the 3 siblings at the time, so it may be a little different.
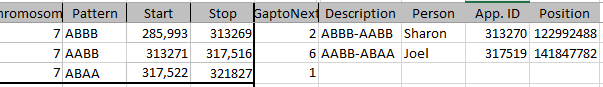
Chromosome 7 Crossovers
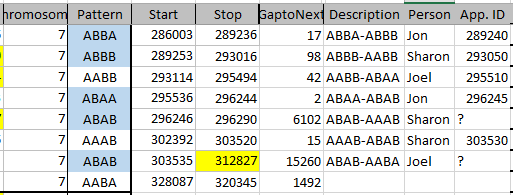
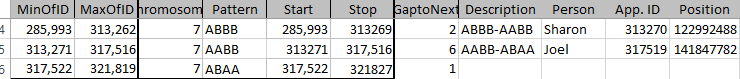
This has been mapped by MacNeill to 3 siblings. Let’s see how my mapping compares. Here is the mom pattern:
Here I have by my own ID’s the start and stop. Then I have gap to the next pattern. This may indicate an AAAA pattern. Under description, I have what the pattern changes are. Then I have the person assigned to the Crossover. Then I have the approximate location of the Crossover. The first line I have the description as ABBA to ABBB. Here, Jon (in the last position) was matching with me as I’m in the first position of course. Then he changed to match with Sharon and Heidi. So I assigned the crossover to him.
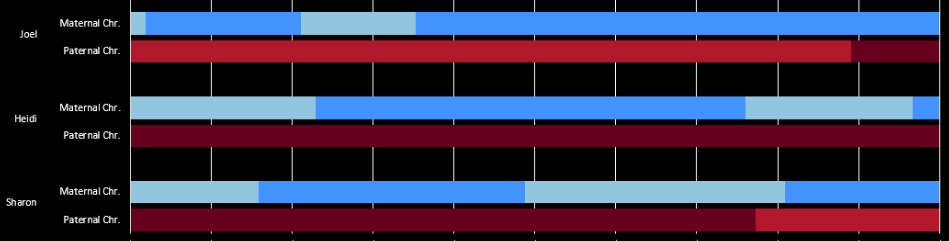
Look at the 5th line. The pattern is ABAB to AAAB. This goes through a gap of over 6,000 ID’s. That usually means there is an AAAA pattern there. AAAA could go to AAAB easily, but to go from ABAB to AAAA would take two crossovers. I don’t have a good idea where the crossover is, so I’ll go to gedmatch. The good news is that I have already tried using visual phasing on this Chromsome:
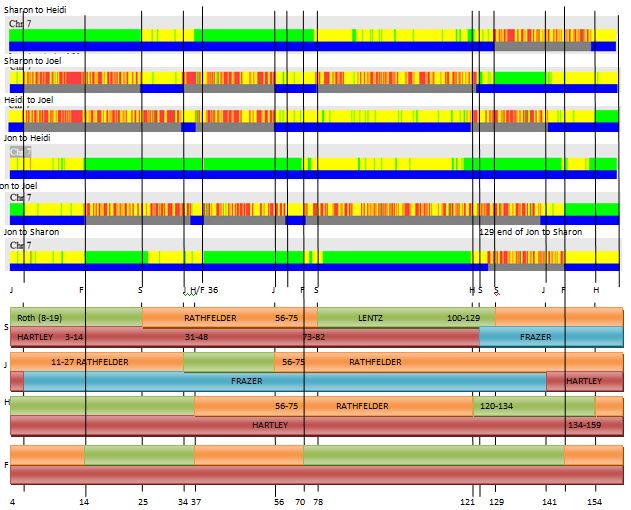
The crossovers that I looked at above in my spreadsheet were on the maternal side. So that would be the top part of the bar (green-orange). It looks like I have 11 or 12 maternal crossovers, if I did it right. Looking at the top part of the image above, notice the non-match areas. These have no blue bar below and have red areas above. These are important. The reason is that if there is any of these areas at any place, there cannot be an AAAA pattern for maternal or paternal. That means that all 4 siblings cannot match the same grandparent in any of these areas. The only potential AAAA patterns, then are at either ends of the Chromosome or in the middle. The middle locations are about 60-70M. Also note that I have Rathfelder as the same match for each sibling from 56-70M.
There is a discrepancy between my spreadsheet crossovers (7) and the visual above (11 or 12). The other problem is that I need a double conversion from my spreadsheet. The spreadsheet is in ID’s which refers to Build 37 locations and Gedmatch is in Build 36.
Before I start converting numbers, I’ll look at what I have for the Dad Crossovers.
Here I added a position number for the Chromosome (Build 37). This matches up with the visual phasing above. What is missing would be the crossover for Joel after an AAAA pattern at the beginning of the Chromosome.
Where is Heidi?
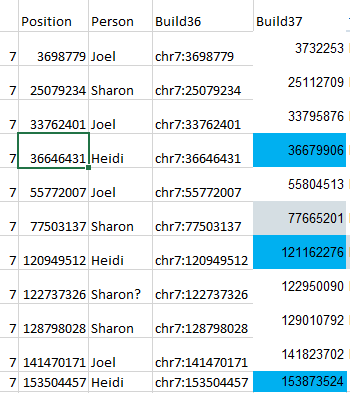
As I look at the maternal visual phasing, I see that Heidi has 3 crossovers. On my spreadsheet, she doesn’t have any. One can be explained as going onto the right end of the Chromosome to an AAAA pattern, but what about the other 2 crossovers, in the middle of the Chromosome? I got these positions from an old file where I compared myself to my 2 sisters. Then I put those in a spreadsheet and converted them to Build 37:
The Chromosome position numbers in blue were where I had Heidi’s crossovers. I then went to my Access Database.
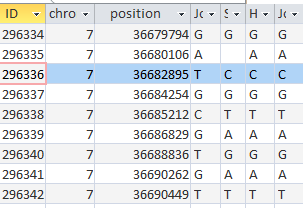
Heidi found
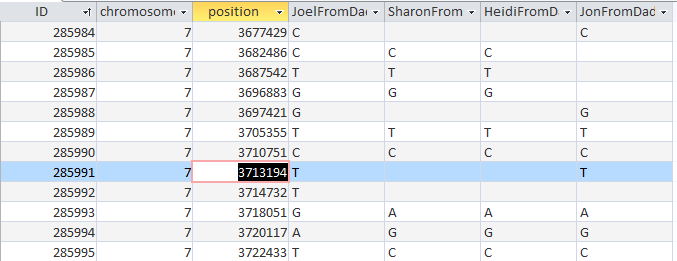
Here is an ABBB Mom Pattern that I missed. Going through the list, I updated my crossover list:
Now I am up to 12 Maternal Crossovers. The AAAA patterns tend to fit in naturally. Note next to the first blue ‘Joel’. There would be no way to go to an ABBB pattern to an AAAB pattern without 2 changes. That is why an AAAA pattern is required within the other 2 patterns.
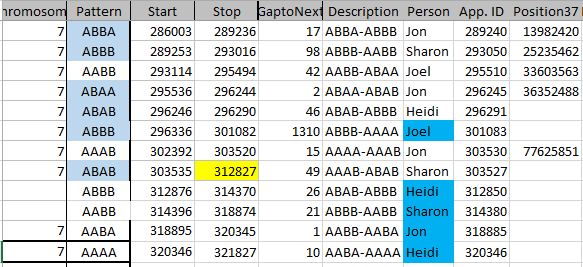
Paternal Crossovers – Chromosome 7
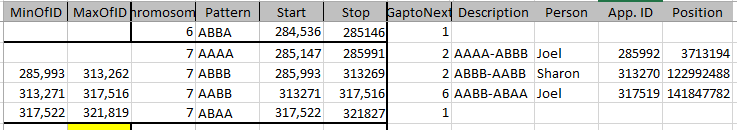
Here I only show 2 crossovers, where on my map above, I show 3. I am just missing my own crossover from AAAA to ABBB. This is at the beginning of Chromosome 7. Here is my database table for my Dad Patterns:
The position I have highlighted would still be an AAAA pattern as I have A??A. So that is the last position with that pattern. Id 285993 is the first spot I have the ABBB pattern, so I chose the crossover as ID# 285992 (under App. ID):
Here is what MacNeill had for 3 siblings at Chromosome 7:
What is now clear from have 4 DNA tested siblings is that my first crossover is paternal and not maternal. For my first crossover to be maternal, I would have had to have gone from an AAAA pattern to an ABBA pattern which would have been a double crossover. Having my brother Jon (the last ‘A’) tested made that clear.
Summary
In this Blog, I have looked at the Mom Patterns created by 4 siblings. Based on those patterns I have filled in alleles from other siblings. I have also filled in alleles for heterozygous siblings. This is based on the Mom allele being known and assigning the other allele as from the Dad. Then I looked at assigning crossovers to the various siblings. Based on the Patterns, it seemed clear who the crossovers should be assigned to. I then checked the crossovers I had with a visual phasing based on gedmatch. This showed where I was missing crossovers, which I was able to add using Chromosome 7 as an example.
Next: How to show the final results?