Homozygosity – Zero to Eighty in One Day
In my previous post, I discussed Homozygosity. In that post, I got my brother Jim from zero phased to 80% phased in one day. Although raw data phasing is considered advanced, the principal of homozygosity is very simple. It just means that you have two alleles at a location that are the same. If your parent has two alleles the same, then you got that allele from that parent’s side. If you have two alleles the same, then you got that allele from both your mother and your father.
Heterozygosity – Two Different Alleles at a Location
Heterozygosity is a little more complicated. It means that you have two different alleles at the same location. Genetics tends to be binary which to me is very simple. Binary is yes or no. You either have XX alleles at a location or XY at a location. A heterozygous results is XY.
Whit Athey and Heterozygosity
Whit Athey has a paper on Raw DNA Phasing here. This is his third principle:
Principle 3 — A final phasing principle is almost trivial, but it is normally not useful because there is usually no way to satisfy its conditions: If a child is heterozygous at a particular SNP, and if it is possible to determine which
parent contributed one of the bases, then the other parent necessarily contributed the other (or alternate) base. This principle will be very useful in the present approach.
Where is Jim Heterozygous?
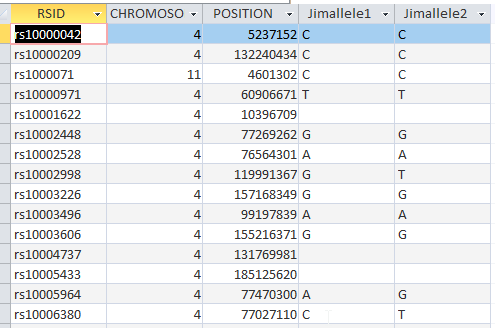
I need to look at Jim’s Raw Data File. I’ll ask Access to find Jim’s alleles that are different:
Jim is heterozygous at a little under 200,000 locations:
Where am I going with this? In the last line above, Jim is AG. If I know mom is A, then Jim has a G from Dad at that location.
Getting Dad Alleles From Mom
In this Query, I am taking all of JIm’s Mom allele’s that have no corresponding Dad alleles. These are the allele’s that he got from his Mom being homozygous. Then I linked those results to Jim’s heterozygous results. That ends up looking like this:
There are over 96,000 locations where we can fill in a Dad allele for Jim. In the first line above, Jim has a C from his Mom. Jim’s results are C and T, so the T has to be from Jim’s Dad.
Putting It All Together – Adding Jim to My Other 5 Siblings
I could figure out how to get the T into the JimFromDad Column above. But I really need to get Jim into the Table I already have with his 5 other siblings and Mom. It would be nice to add Mom’s FTDNA results to that table also. Right now that Table has 26 columns and I want to add more.
Here is the structure of the existing 5 sibling table:
I wasn’t too consistent on my capitalization. The sibling Dad alleles are grouped together as are the sibling Mom alleles. This is for comparison. These sets of Mom and Dad alleles will form a pattern that will determine the crossovers. The above table is called tbl5SibsHeteroMomtoDad, so it is at about the same stage that I am with Jim.
I’ll try this query to add in Jim’s Alleles 1 and 2:
Here I made an unequal join, but I don’t think that will work. I want everything from Jim’s list and everything I already had in the 5 sibling list. This will probably call for an Append Query.
In order to perform an Append Query, I need to have the same column headers. I copied the 5 sibling table and pasted it as a six sibling hetero mom to dad table. Then I added some columns for Jim:
I’ll also add some Mom and Dad allele columns for Jim. Next I open up Jim’s original download table into a Query:
I select Append at the top and choose the Table I want the data to be appended to:
I choose View to see what I have and it shows 720,449 records which sounds right. Then I choose Run.
This didn’t get me what I wanted. It added an extra row for Jim. When I sort by RSID, it looks like this:
It is giving Jim an extra row for his results, which I don’t want. Abort this mission.
A Right Join and a Left Join?
I can go back to my original thought. However, it will take two steps.
First I want to note that there are 942,647 rows in the 5 Sibling Table. There are 720,449 in Jim’s raw data table. I don’t want to lose any data along the way. I put an ‘is not null’ into Jim’s allele 1 column and got 720449 rows of data, so one query was enough. I like this so much, I’ll make a Table out of it:
This didn’t work so I tried repairing and compacting the Access database again. That seemed to solve the problem.
Now I have a new six sibling table. For five siblings, I have the Mom and Dad alleles of the first three steps. For Jim, I just have his raw data included so far.
Mom Vs Mom – Ancestry and FTDNA Results
Now I am wondering if I need to add Mom’s FTDNA raw DNA data to my table. Mom has 701,478 rows or positions at AncestryDNA. Mom has 711,398 rows at FTDNA. That is about 9,000 rows difference, so I guess it is worth it. It could make the Table more complicated for comparisons. If I can can combine, mom’s alleles into two columns instead of four, that would be better.
Here is my comparison of FTDNA vs. AncestryDNA for my mom:
This query will return all the RSID’s that are in FTDNA but not at AncestryDNA:
That is over 23,000 results. I will need these, if I am to recreate Jim’s results.
Getting Mom’s Extra FTDNA Results into My Six Sibling Table
First, I created a Query to find out how many RSIDs mom had from FTDNA that were not already in the six sibling table:
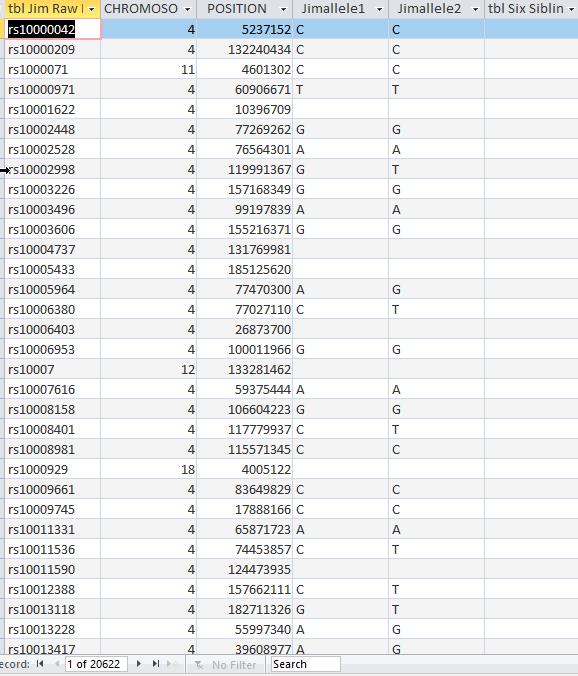
This tells me Mom has 17,935 positions tested that are not in the Six Sibling Table. However, if those are positions that Jim has, I will want to add those also. I checked and Jim has 17,835 positions tested out of mom’s 17,935. I was curious, so I checked Jim’s positions that weren’t in the Six Sibling Table and got 20,622. These are the details that bog me down.
Appending to the Six Sibling Table
I want a good Six Sibling Table, so I’ll append the 20,000 positions that Jim has that are not in the table.
Here is my Append Query:
The Query says to add only Jim’s raw DNA data to the Six Sibling Table that isn’t already there. When I view what is to be appended, I get the right amount of rows.
When I hit the Run button, I get this error:
I was wondering if that would be a problem. I don’t want the extra rsid column. I need to save the underlying Query first. I did that and had the same problem, so I made a table out of the Query.
That looks better. I think this will work:
This worked. So now my master table has 963,269 rows. Bigger is better as long as it is good data.
My plan was to add my mom’s alleles, but so far I have only added Jim’s. When I now check the positions that Mom has that are not in the Six Sibling Master Table, I only get 100. There are actually only 99 extra rows as one was a header that I deleted:
I’ll follow the same procedure. I’ll make a small table for Mom and then append it. I’m not sure of the significance, as Mom may have no siblings corresponding to these alleles at this time.
Here is the new Master Table with Mom and Jim appended:
One More Master Table Adjustment
On the Six Sibling Master Table I added a place for Jim Dad and Mom alleles:
I probably should have done this before I phased Jim. However, the advantage is that I have Jim’s results separate from this table that I can check on. I can now re-do the processes to get Jim’s phased alleles or try to copy what I had into this master table. [Note I try to copy Jim’s results below, but the results are not good, so I end up recreating his results in the Master File that has the results of all six siblings in it. See section called Plan B below.]
I’ll try to use an Update Query to get Jim’s phased alleles into this master table. Here is my Google search for Update Query:
Actually I thought of an easier way:
Here I took the whoe Six Sibling Table and replaced Jim’s phased alleles where he had none. I only get one shot at this, so before I do this, I’ll add Jim’s heterozygous phased alleles to his two homozygous alleles.
An Append Query for All of Jim’s Phased Alleles
I appended Jim’s Heterozygous phased alleles to his homozygous phased alleles.
Here is the point at which Jim’s phased alleles are based on what he got from his mom to what he got from his dad. There are only two problems:
- The name of the Table is now wrong, so I need to change it;
- I never added in the alleles that Jim got from Dad. That is OK as I have the information to do it.
Adding Jim’s Heterozygous Dad Alleles Based on Mom’s Results
Now I am back to where I was before I took a detour of incorporating Jim and my Mom’s FTDNA results into my existing five sibling table.
Here I’m going to cheat a little and look to see what I did in the past:
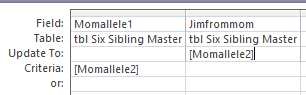
Here’s my sister Lori. Back when I knew more what I was doing, I had an Update Query whcih said, ‘When Lori’s allele 1 was not the same as her allele 2 [heterozygous] and Lori had allele 1 from mom, put Lori’s allele 2 in her Dad spot’. Seems like that should work for Jim.
When I press View, I didn’t get any results. I have a guess as to the reason. This may have to do with the situations where Jim got his Mom allele and he had no results for himself. I tried i this different ways and could not get this to work unless I took out the expression: <>[Jimallele1].
This makes me think that something is wrong with the Table. I checked for duplicates in the table and got 96,222.
So this is good to know. At rs1000002 there are two results. One has Jimallele 1 and 2 rfesults and one does not. However, at rs10000300 there are no Jim alleles and there is only one result.
Plan B – Work with the Six Sibling Master Chart
I checked the Six Sibling Master Table for duplicates and didn’t find any. I think I’ll just work with that Table.
Here is Step one from Whit Athey:
Principle 1 — If a person is “homozygous” at a location—that is, having the same base on each of the two chromosomes of a pair, then obviously at that location it is possible to know with certainty that both chromosomes of the pair have that base at that location, but this is an almost trivial form of phasing.
I had a little practice trying to get the Update Query to work. Now I’ll try it on Jim’s results in the Master File. Unfortunately, I am still getting no results. I decided to go ahead and run the Update Query even though I saw no results in the View mode. This was after making a backup up the Six Sibling Master File. It looks like the update worked.
The Update Query was quite simple. It said if Jim’s allele 1 and 2 were the same, then give that allele to his Dad and Mom side.
Updating Mom’s Homozygous Alleles to Jim
The next Update Query will be similar:
This says if Momallele 1 and 2 are the same, give Jim one of those on his maternal side. Here is the warning:
Here are some of the results.
I hope to catch the blue line in the next query.
Updating Jim’s Heterozygous alleles where Jim has a Mom allele.
This Update Query says when Jim is heterozygous and he already has his allele 1 in the mom spot, put allele 2 in the Dad spot.
I am down to a mere 34,000 rows on this Update.
Next, I want to switch the alleles:
When Jim’s allele 2 is in the Mom place put Jim’s allele 1 in the Dad place. That should fill in these blanks:
Here is a summary of what I have for phased alleles for me and my siblings:
One interesting thing is that Jon has 751,171 maternally phased alleles. Jon only tested at 668,942 positions. The additional results must be where Mom had results at positions that Jon didn’t test at. That is assuming that I didn’t mess up somewhere.
One More Query for Fun
This is looking for childrens’ missing alleles from Mom when Mom has two alleles that are the same. I found a few:
These are likely positions that were not tested by my siblings. I made a quick Update Query to add those Mom alleles in for my siblings.
Summary and Conclusions
- I started out looking at my brother JIm’s heterozygosity. I found out where he could have an allele assigned to his paternal side in the case where we knew his materal allele.
- I worked on getting Jim’s results into the master file I have for his other 5 siblings. I also added some more of my mom’s alleles that were from FTDNA and not included in her previous AncestryDNA resutls.
- I tried to get JIm’s alleles phased before I brought them into the five sibling file, but I ended up with duplicate results.
- I decided to work with the Six sibling file which had no duplicates and recreate Jim’s phased alleles based on the principles of homozygosity and heterozygosity. I was able to do this quickly with Access Update Queries.
- I now have a large master file with 30 columns. These columns have the raw data for my mom and her six children as well as their alleles that have been phased so far. I will be working with the last 12 columns in the upcoming Blogs. These are the patrernally and maternally phased alleles. They will form patterns that will tell me where the crossovers are.