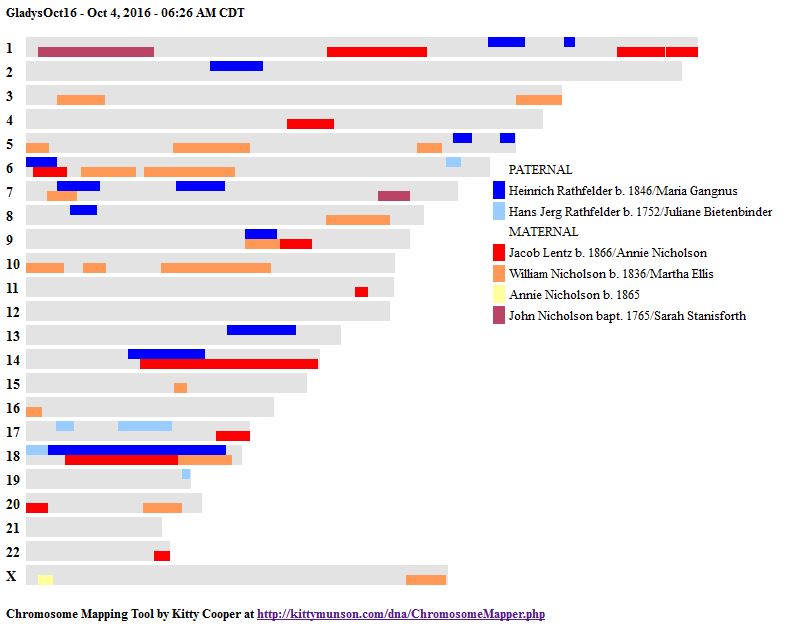
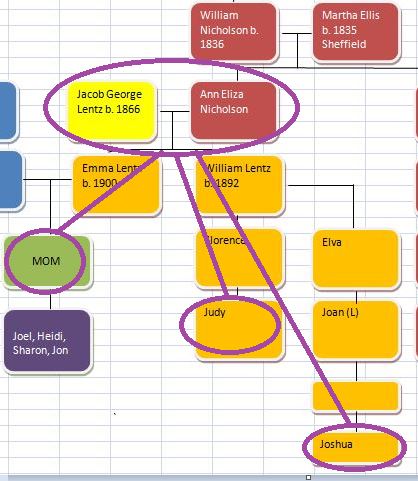
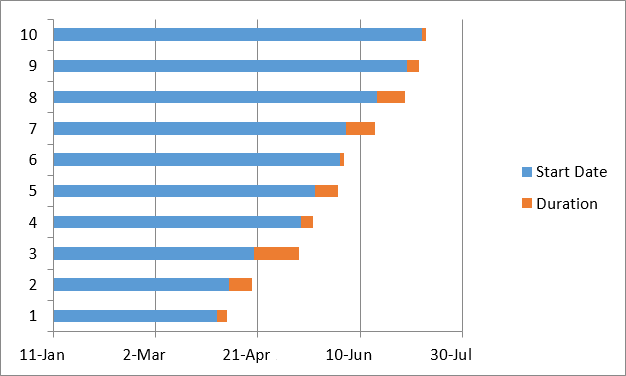In my past 4 Blogs on the topic, I have started the phasing of my siblings raw DNA using my mother’s raw DNA. I used Whit Athey’s applicable paper on the subject, MS Access and have checked my results with the work that M MacNeill’s similar analysis of my raw DNA. I started out using 3 siblings in the analysis. Part way through, my brother’s results came in, so now I am looking at 4 siblings.
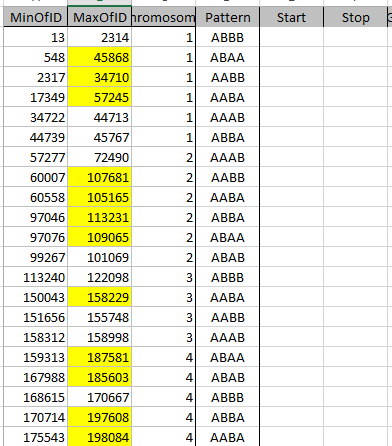
I parted a bit with the Whit Athey analysis in that where he went to a visual analysis, I decided to look for the change points in the data. I then used those points to perform an Access query to update the various patterns found. When I left off my last Blog, I had just located Starts and Stops for the 7 different paternal patterns for the 4 siblings.
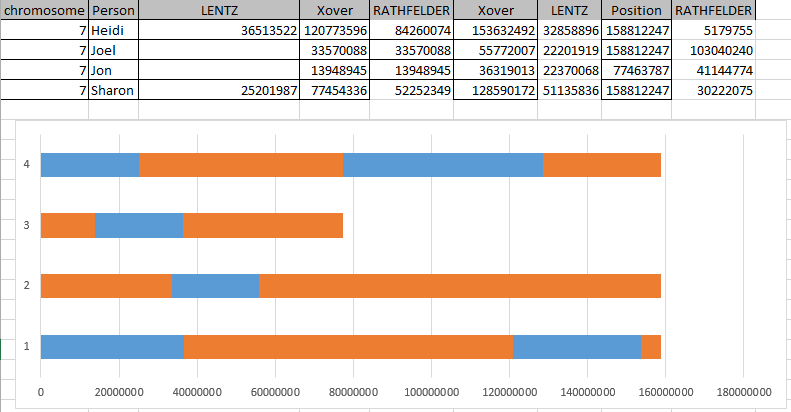
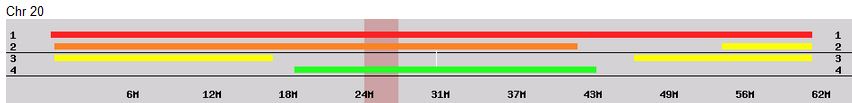
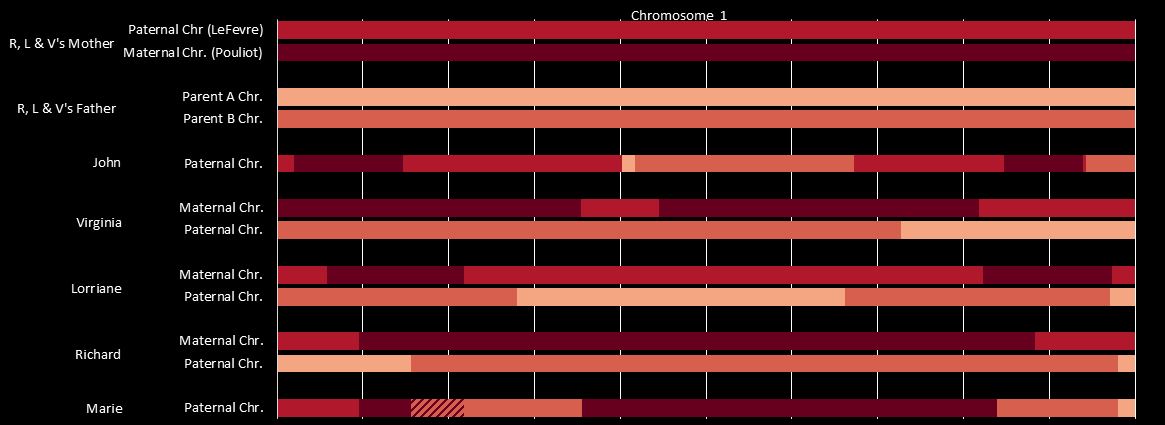
Sibling Patterns in an Excel Stacked Bar Chart
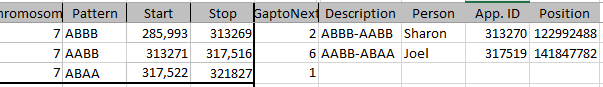
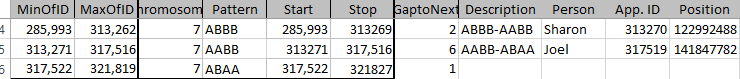
Today I was getting a headache trying to find a way to put the paternal patterns information into Excel in a Bar Chart. Here is the best I could do for the first 2 Chromosomes:

The spreadsheet data format is above. I chose Stacked Bar Chart. Then I had to transpose the row and columns. The slight glitch is that I had to create an extra duplicate pattern when that occurred to get the results in one bar per chromosome. I used the end point for each pattern. The bar assumes the start is at zero for each chromosome which isn’t totally accurate, but close enough, I suppose for a bar chart. The bar chart is meant to represent all the paternal changes in patterns for me and my 3 siblings.
When I check the change in patterns to the number of crossovers in the work of M MacNeill, it appears that I have missed a pattern change on each of the above crossovers. Hopefully, I will find them as I go through the process and re-check my work. I guess I’m batting 2 out of 3 now.
Finding the Two Missed Paternal Crossovers
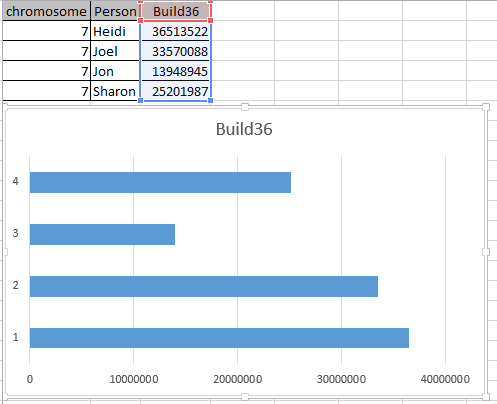
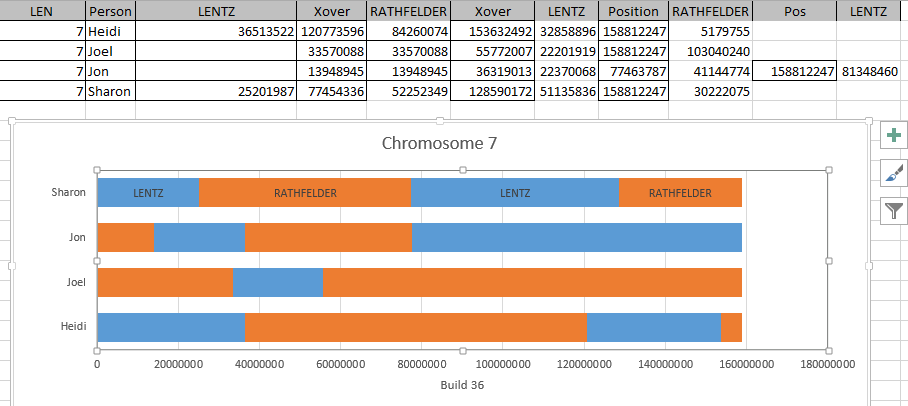
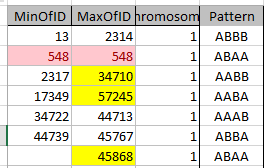
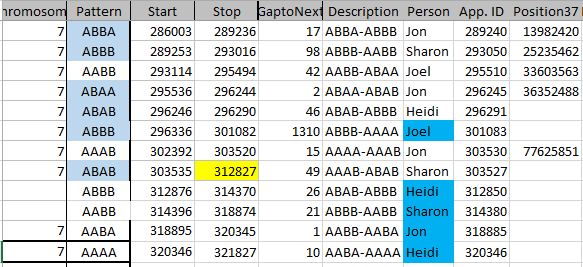
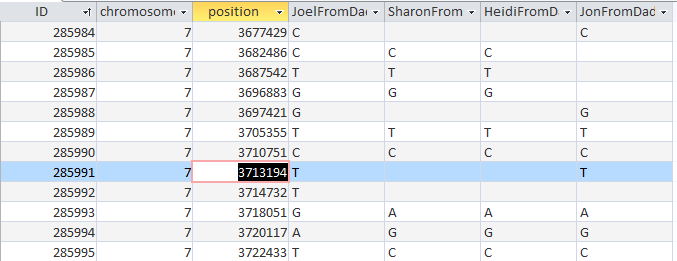
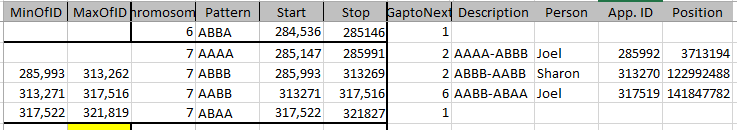
It is possible that they aren’t missing at all. Perhaps in all the work I did to represent the information in an Excel Chart above, I misrepresented the work I had done. Here is my spreadsheet for Chromosome 1:

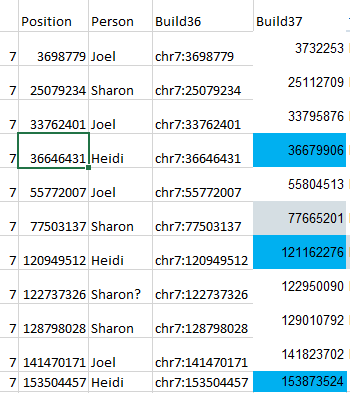
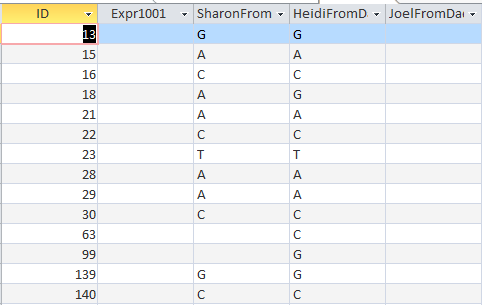
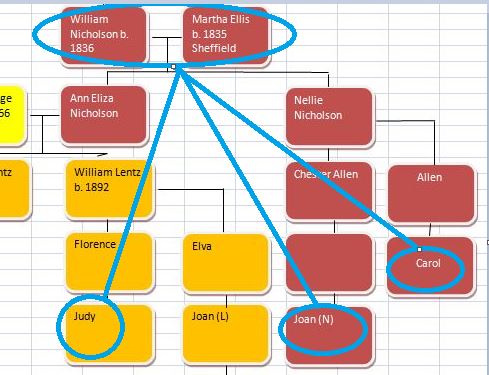
Here is what M MacNeill has for Joel, Heidi and Sharon’s paternal Starts and Stops on Chromosome 1:

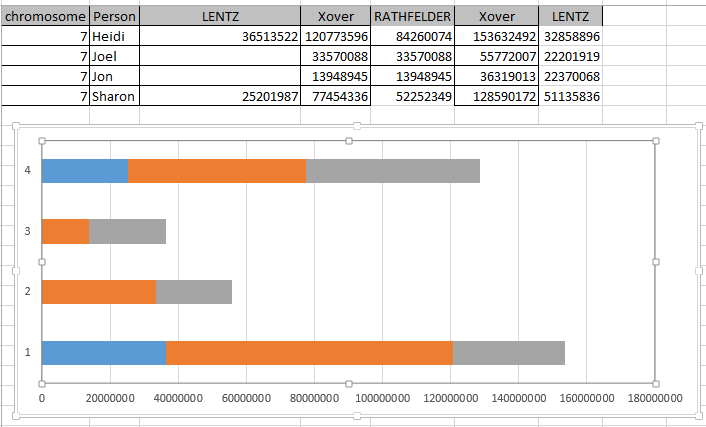
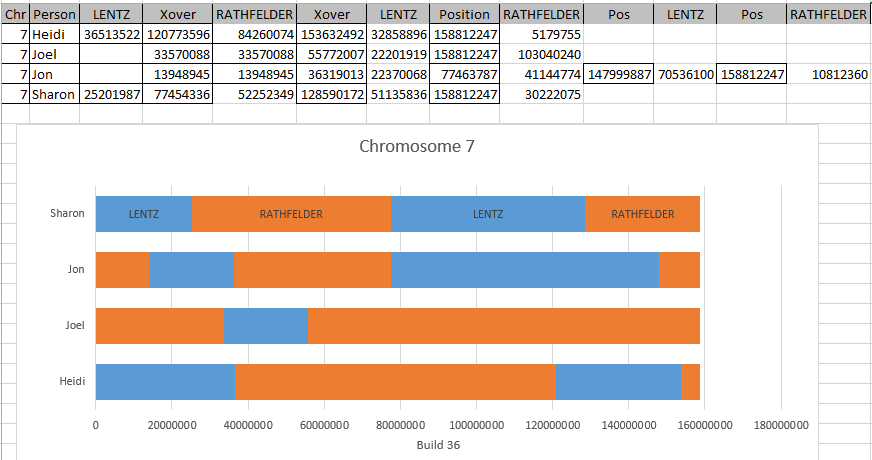
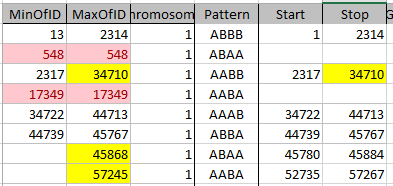
It looks like MacNeill has 6 paternal starts and stops. I don’t count the last one as that goes to the end of the Chromosome. Again, I run into the conversion issue between my Build 37 and MacNeill’s Build 36 work. Here is what happen when I put the approximate crossover locations side by side:

This shows that we both have 6 crossover, which is good. It gets a bit confusing. Note that I had to add a crossover at my position 23289397. That is because there was a gap. That is the gap where the 4 siblings must match the same paternal grandparent. Normally, there shouldn’t be any gap between the Stop from one pattern and the Start of the next one. So it turns out I’m doing better than I thought. That is encouraging to know. For the last pattern, I don’t have an entry, because the crossover is at the same spot as the Stop of the previous pattern.
However, I am comparing my 4 sibling work to MacNeill’s 3 sibling. Also MacNeill had a start of 742429 and mine was higher. That means that there must be a pattern between 742,429 and 1,062,638. I checked, and there aren’t many extra locations there. I suppose I did as well as I could do. I do wonder where Jon’s Chromosome 1 crossovers are, though. Perhaps he has a double crossover with another sibling or one that is very close in location to another sibling.
Gedmatch Check
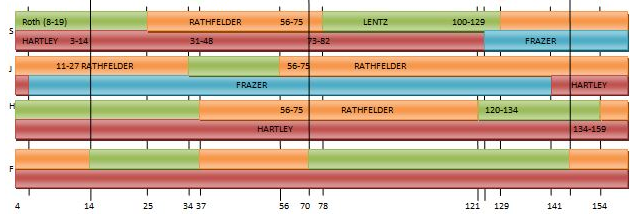
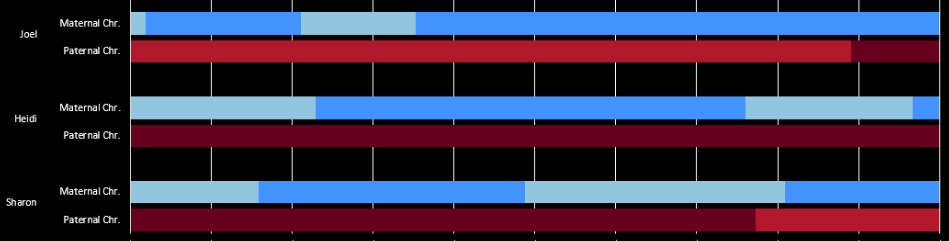
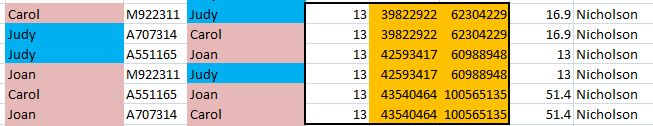
Here is how my 3 tested siblings and I look compared to each other at Gedmatch in the browser:

The lines don’t match up perfectly, but I have for 3 crossovers for Sharon in a row. I am J, my brother Jon is F and only has one crossover of his own. These are the combined paternal/maternal crossovers. When I map it out using a visual method, it appears that Jon may have no recombination in Chromosome 1.

If that is true, it would make him a good candidate for finding Frazer or Lentz relatives at Chromosome 1.
assigning Paternal crossovers for Chromosome 1
Assigning crossovers is getting a little ahead of myself, but I would like to see if I am on the right track.

Here the Dad4Pattern represents Joel, Sharon, Heidi and Jon. There appears to be a logic to assigning these crossovers that I have in the XSib column. The first crossover I have going to Sharon. That is between the first Stop and the second Start. Sharon’s B in ABAA goes to an A in the AAAA homozygous region. That means all siblings match the same paternal grandparent in this region. The next crossover goes to me as I’m represented by the A in the ABBB pattern. The 3 other siblings remain matched to each other. Then Heidi gets the next crossover as she goes from matching Sharon and Jon to matching me. The next to the last crossover, I had as Jon. But it has to be Heidi as she went from matching me and Sharon to matching Jon. If Jon was the one that changed to matching Heidi, the pattern would have gone to AAAA. Likewise, the last crossover I had as Heidi, but it has to be Joel. I went from matching Sharon to matching Heidi and Jon.
There are a few cross checks to the method. One is to check to see what MacNeill has done. Another way is to check to see known matches. I noticed above that Jon had matches to known matches on my paternal grandmother on either side of the wrong crossover that I had assigned to him, so that was likely not a good crossover. Another note is that there is a least one other homozygous region. That is between the 191M stop and 192M start above. That means that there should be an AAAA pattern stuck in there, but it is not necessary to know at this stage.
Time to Push the Button: Back to Phasing by MS Access
A lot of the above work was to make sure that I had the right number of crossovers in the right places. I was worried that if I didn’t, that I wouldn’t be applying the right rules to the right areas of the spreadsheets.
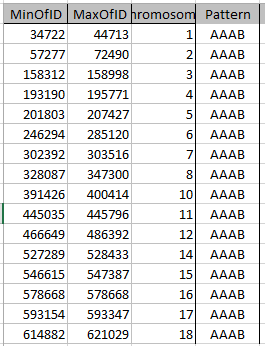
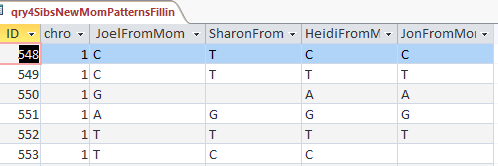
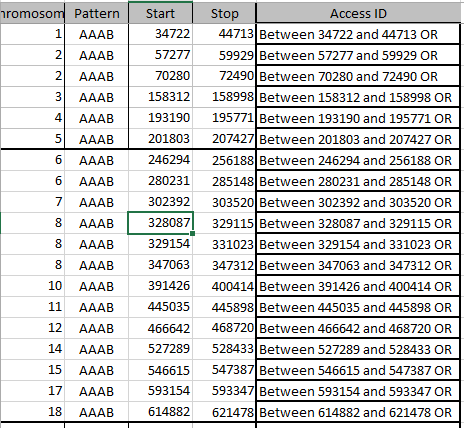
First aAAB
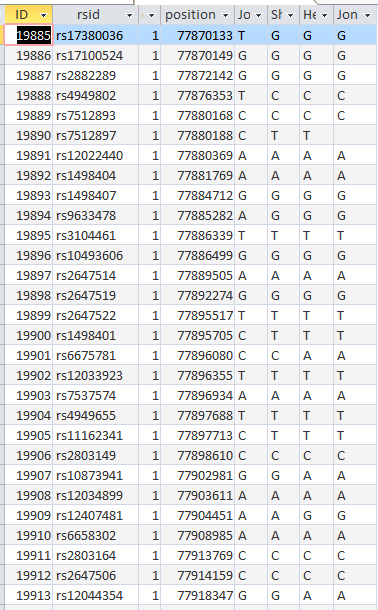
Here are my AAAB paternal Patterns with start and stop in Access language:

Here are some examples of fixes that are needed within these AAAB areas:

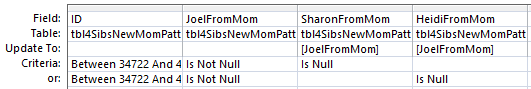
Basically, if there is a blank in the first 3 positions, it should be filled in by the non-blank in that area. But how do I write that into a formula? Here is one way:

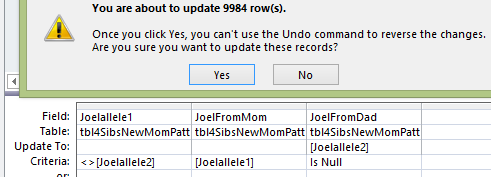
This says if Joel’s Dad base or Heidi’s Dad base is null, put Sharon’s value in. I ran that and it updated 2165 rows.
Next:

This time only about 1400 rows were updated. The last time we fill in Heidi’s value if Joel or Sharon had a missing Dad base.

I’ll check my work. I see a flaw in my logic already. I shouldn’t have put the 2 ‘Is not null’s’ on the same line and Access sees that as an and. I wanted an ‘OR’, so they should’ve been on separate lines. Here is the revised query putting Heidi’s Dad base in the empty spot of Sharon and Joel.

Note that I had to put the position criteria for the Paternal Patterns in twice also. See, I had missed 4121 rows. I went through this with the 2 other siblings.
AAbA Paternal pattern

The AABA also has potential for filling in.

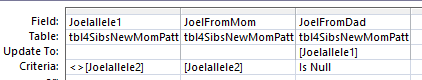
In the first line there is ?T??. In and AABA pattern, we know that the first and last position will also need to be T. In the second line, we don’t know what to fill in. In the 3rd line we can put a C in the last column.
My ID locations for AABA look like this:

The queries will be similar to last time except that they will involve Joel, Sharon and Jon and leave out Heidi.

This was a more popular fill-in. In the query above, if I had a Dad Base and Sharon and Jon didn’t it went to Sharon or Jon. I then did the same thing for Sharon and Jon. Here I check my results.

These are the same ID Lines shown as above before I did the query. This now shows that Joel, Sharon and Jon have the same bases for this AABA Pattern. This is even true when we don’t know the base Heidi has on her paternal side as in the first row.
aabb pattern

Here is what an AABB Pattern area looks like before I fill in the bases:

The rule is if Joel or Sharon or Heidi or Jon have one base and the other is missing, fill in the missing one with the one that is there. However, as in the second row, where Heidi and Jon are both missing, nothing may be filled in. This will take a little thought. Perhaps I can do this in 2 steps:

This says if I have a base AND Sharon doesn’t, give her my base, then also do the same and fill in Heidi’s base to Jon’s missing base from dad. This query filled in a little less than 20,000 bases with the push of a Run button. Then I’ll do the opposite:

This time Sharon’s base goes to me and Jon’s goes to Heidi. I’ll check good old ID 45494.

It looks like I filled in what I wanted to and didn’t fill in what should not have been filled in.
The other combinations will be variations on what has just been done. Either 3 will match each other and one won’t or there will be 2 pairs that match each other within the pair.
abaa Fill-in
This is the first pattern of my siblings’ 1st Chromosome.

Another ho-hum 20,000 rows filled in.
Here Heidi fills in Joel and Jon:

The updated rows go down the 3rd time I run this.

abab Dad pattern
This will be where Joel and Heidi match paternally and Sharon and Jon match.

Jon is probably missing a lot of bases due to being tested on with the Ancestry Version 2.

abba

This query says in the ID areas where there is an ABBA pattern put Jon’s dad base into Joel’s missing dad base area and put Heidi’s dad base into Sharon’s missing dad base area in the table called tbl4SibsPPatternFillin.

Here, I made a mistake. Note that I had Access overwrite a bracket “]” that didn’t get erased. That means that I will have to run this query again to get my bases back from Jon. Here is what the above Update Query did.

Fortunately, Jon still has the bases that I gave him. I’ll redo the query to get my bases back.

This query will fix my error. It says if I have an end bracket as a base, fill it up with what Jon has.

abbb – the last paternal combo
This time I won’t touch my bases, but make sure that Sharon, Heidi and Jon match.



So that should have filled in all the paternal patterns.
Finding the AAAA ‘Patterns’
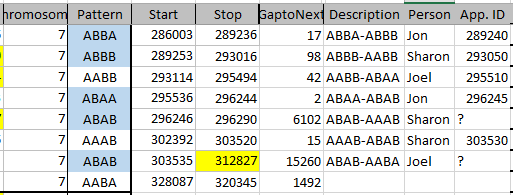
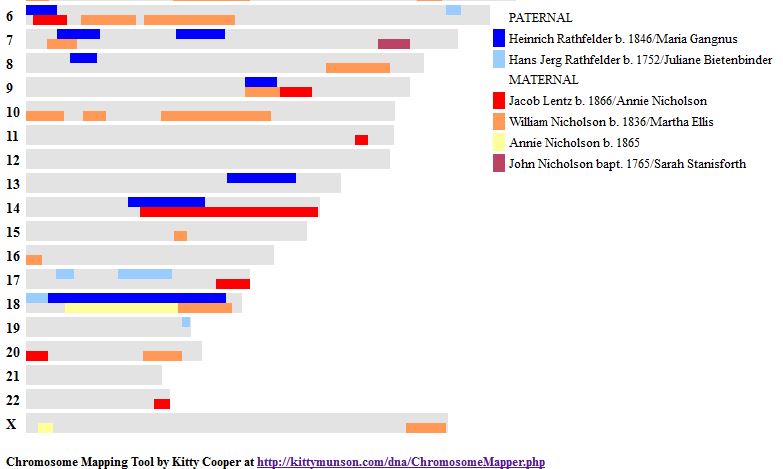
This should be a little trickier. Previously, we had identified one AAAA pattern in Chromosome 1. This can be seen between 19 and 23 below. All the paternal areas are orange.

There is no other area on this Chromosome that is all orange or all green for all siblings. However, how do I identify all the other quadruple A patterns? It is not as easy as the other patterns because this pattern may occur within other patterns. I could make a chromosome map for each chromosome as above, however, it becomes a chicken and egg problem. It would be nice to know the AAAA areas so I could draw the map.
Here is a spreadsheet where I checked the number of IDs from the Start to the previous Stop.

When the amount was more that 500 IDs, I highlighted that number in yellow. Above between the Stop of ABAA and the Start of ABBB on Chromosome 1, there was an AAAA pattern for 1478 position numbers.
The next yellow area is in Chromosome 2 which is a larger region of AAAA
pattern.
Here is an interesting situation:

This yellow area is above the amount I chose as a minimum of 500 positions. However, as I look at my worksheet, I see that the ABBA pattern extends beyond ID# 285124. So I will do a new query based on the new fill-in table. Here is the new ABBA:


This shows that the ABBA pattern goes to the end of Chromosome 6. I can fill in the extra letters by hand and adjust my spreadsheet.
However, what about Chromosome 7?
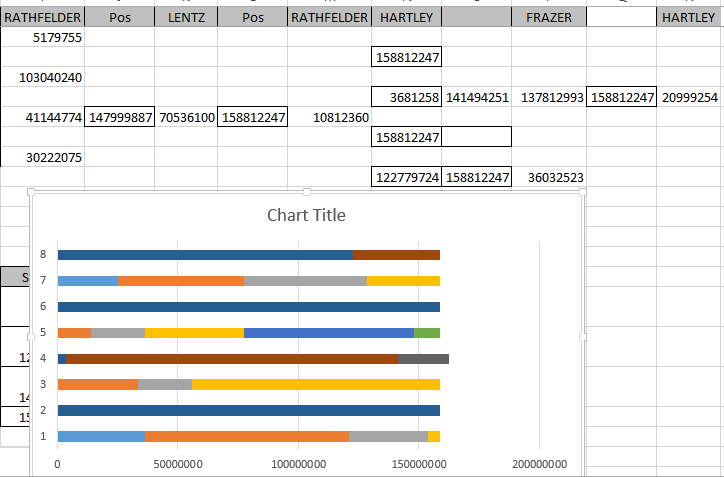
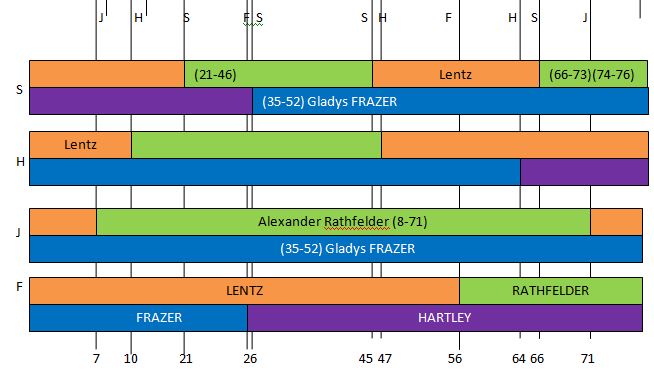
Chromosome 7 appears to have an AAAA pattern for about 847 ID#’s. This is how MacNeill mapped my Chromosome 7.

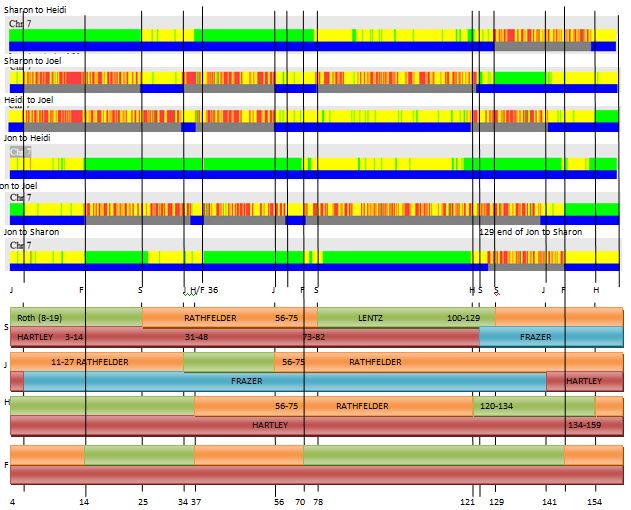
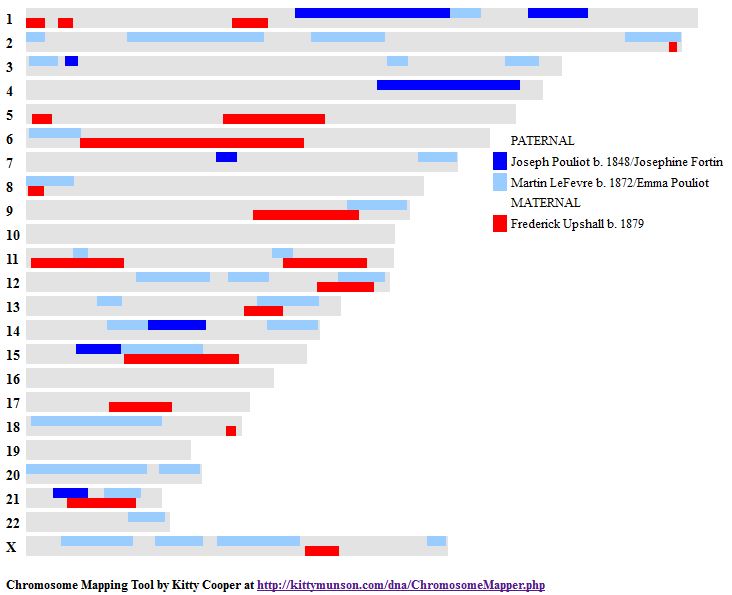
He would have the ABBB Paternal Pattern with me being the A. This is how I had visually mapped Chromosome 7:

These end pieces are difficult where there is a half identical region. I will stick with my as I do notice a small match with my Hartley-related 2nd cousin Pat:

This may become more clear once my brother Jon is mapped out. In fact Jon is Fully Identical with me in that region:

Jon also matches cousin Pat in that same spot:

Ergo, I must match Pat aka Hartley DNA at the first part of Chromosome 7.
Here is Jon mapped out no Chromosome 7:

Jon (F) and Heidi (H) got a full dose of Hartley DNA at Chromosome 7.
That was a bit of a long exercise, but the intention was to prove to myself that an AAAA pattern of over 500 positions (or my ID#s) is a valid AAAA Pattern.
Filling in the aaaa’s
As I have now convinced myself that this small area was indeed an AAAA area, I can proceed. I made a formula in Excel that takes the other Patterns’ Stops and Starts and puts them into Access language.

The formula adds an ID# to the beginning and subtracts one from the end so the AAAA patterns have their own range.
Inspecting my work
Having found a pattern boundary that was off at the end of Chromosome 6, I will check the other boundaries. According to my spreadsheet, the first AAAA should end at 6604.

The actual Access Data table is different by one:

That mean that the I need to add an ‘A’ to the missing space and change the start of the ABBB Pattern from 6605 to 6604 – a pretty minor change. I made a few more minor changes. However, I’ll hold off on making the AAAA pattern changes for now. That is in case the boundary changes again due to other changes I’ll be making.
Filling In Mom Bases From Dad Bases
This is about how far I got last time when I was trying to phase 3 siblings. My interpretation of this portion of the process is to look at the heterozygous siblings. Where they have a new base on the Dad side, they will know that the other base goes on the Mom side.
Finding heterozygous siblings
First I made a new table to put the new information in. It is just a copy of my last table of the fill-ins based on patterns. Here is a query just to find the alleles for each sibling that are different from each other:

Here is the Update Query. I better get it right as it is doing a lot of things:

The first part has the criteria that makes a person heterozygous. I forgot to make sure that the mom base was missing, so I need to add an ‘is null’ phrase:

This may not be necessary, but just makes sure I am not overwriting anything that is already there. So when mom’s base is missing add the base that isn’t the dad base. Or more specifically, add allele2. This changes 39,260 rows.
Next to get the opposite effect, I change most of the alleles 1’s to 2’s and the 2’s to 1’s.

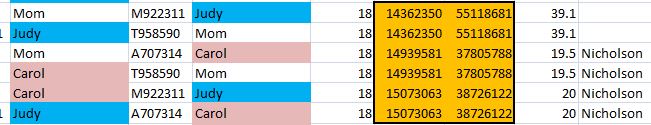
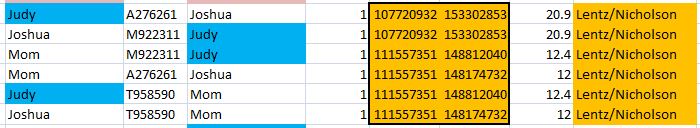
That changed over 10% of all the results. To check, here is a query from the older un-updated table showing just my results where I’m heterozygous and my allele1 was from Dad:

Here is the updated table.
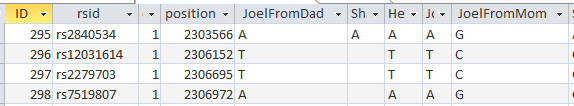
The G, C, C, G was added as a base from my mom – along with 10’s of thousands of other bases.
Summary
In overview:
- Principal 1: I’ve added the homozygous sibling results. This says a double base for a sibling means that they got the same base for each parent.
- Principal 2: I forgot to add the homozygous mom results to Jon. I’ll do that in the next Blog
- Principal 3: This is for heterozygous siblings. When one base is known for a parent and the other parent base is missing, the other base is assigned to the other parent
- Next I looked at the paternal patterns and made note of where they changed
- For each paternal pattern region I filled in the bases that could be filled in based on that pattern
- Then based on that new information, I filled in more missing mom bases from the dad bases in areas where the children were homozygous. This is Principal 3 reapplied.